Tips and tricks for Galaxy
My years of experience working with the Galaxy analysis platform has yielded a useful bag of tricks and tools that I would like to share. This is intended to be an overview more than a tutorial, but please don’t hesitate to contact me if you have anything you would like to add or is unclear. I hope you find something useful here.
Intro to Galaxy
The Galaxy Project aims to bring together the various disparate data science resources and provide a means for public compute resources to host them. Given the sheer variety of infrastructure designs and data storage mechanisms, this was no small task. At its heart it provides an abstraction layer for various data processing and analysis tools. It uses this abstraction to allow chaining these tools into data pipelines (it refers to them as “ workflows”). There are a number of other solutions available to do this task such as SnakeMake, NextFlow , CWL, WDL, and a smattering of others.
Galaxy is distinct because it provides a daemon service to allow the resources to be accessed over the web. It also combines this with data storage, access control, and visualization. One of the most important distinguishing features in my mind is that Galaxy stores its pipelines as a graph data structure. This is important because you invest tremendous amounts of time and money building these pipelines. You want to ensure that that investment is secure against changes in the software over time. Storing as a data structure means that the pipeline can readily be queried and transformed by any software that can read that structure format (JSON/YAML). The other mentioned solutions store their pipeline descriptions as code. This means that you would have to either write a complex syntax parser to transform the pipeline to another solutions format, or manually reimplement the pipeline. Manually reimplementing the pipeline is always going to be plagued with mistakes and failure to capture subtle or implicit functionality.
If you would like to play with Galaxy, one of a number of freely available instances is usegalaxy.org
Intro to collections
One of the most potent features of Galaxy’s workflow functionality is its concept of a dataset collection (list). The
concept of a list of datasets is not particularly revolutionary, but Galaxy’s behavior when working with them is. The
mundane use is for tools to require a collection of datasets to operate on. This requirement is specified in the tools
wrapper and Galaxy ensures that the tool gets what it wants. Galaxy extends this to allow describing collections of
pairs of datasets (list:pair) which is tremendously important when a tool wants to operate on pairs of related datasets
such as what is output from double ended DNA sequencing.
 Collections can be extended even further to allow for collections of collections, which allows you to maintain subgroups
of collections within a collection (list:list or list:list:list). It is important to know that collections are
immutable, once created you can not modify their contents.
They are also more like ordered mappings or dictionaries, they have a unique key that is assigned to each collection element.
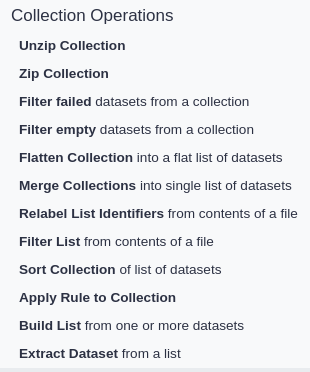
Operations are available to sort, modify, and relabel collections, but they all generate a new copy of the collection
rather than modify them. Most Galaxy instances provide these operations under the “Collection Operations”
section of its tool panel. It is beneficial to become familiar with all of them while setting out to create a workflow.
Collections can be extended even further to allow for collections of collections, which allows you to maintain subgroups
of collections within a collection (list:list or list:list:list). It is important to know that collections are
immutable, once created you can not modify their contents.
They are also more like ordered mappings or dictionaries, they have a unique key that is assigned to each collection element.
Operations are available to sort, modify, and relabel collections, but they all generate a new copy of the collection
rather than modify them. Most Galaxy instances provide these operations under the “Collection Operations”
section of its tool panel. It is beneficial to become familiar with all of them while setting out to create a workflow.
The real magic is when you try to pass a collection of various types to a tool that is not expecting a collection, or collection of collections. This magic is referred to as “mapping over” a collection to a tool. If you pass a collection to a tool that is expecting a single dataset, Galaxy will map over that tool repeating its execution for each collection element. If a tool is mapped over a collection and normally produces a single dataset output, it will produce a collection of outputs. The collection of outputs will be ordered with respect to the order of the input collection.
Advanced mapping
This behavior of mapping over collections goes even further, to where a tool that is expecting a collection can be mapped over by passing it a collection of collections (list:list). You can expect similar behavior when working with a collection of collections of pairs (list:list:pair).
Things get fascinating when you consider how Galaxy behaves when a tool has multiple inputs, and it is being mapped over. If one input is being mapped over, and another is a single dataset, each invocation will receive that single dataset. If both inputs are being mapped over, it is important to ensure that the collections are of equal size or Galaxy will throw errors. The two collections will then be “zipped” (similar to python zip) together, where each tool invocation will receive the nth dataset (or subcollection) from each collection.
One of the most powerful properties of mapping over appears when you consider the simplest collection, the empty collection. What happens when a tool is mapped over an empty collection? It is never executed and results in another empty collection. Keep this in mind as you read on.
GNU Awk in all its glory
Awk, the predecessor of GNU Awk, was originally written in 1977 at AT&T Bell Laboratories. GNU Awk (or gawk) is a powerful little rule based text processing tool. It has a complete scripting language built in that can do everything from generating, reformatting, or filtering text to fully interactive applications than can make network requests. See GNU AWK Users Guide for more information on the software and its applications.
Unlike larger scripting languages such as Python, gawk remains simple enough that it can be effectively sandboxed. This allows it to be provided as a public resource for use in data analysis pipelines. I spent some time ensuring that I could not find a way to break out of the sandbox, and despite its age, actually managed to find two significant vulnerabilities that could provide attack vectors to its host system. Arnold Robbins, one of the authors of gawk, was quick to respond to my reports of these issues and version 5.0.1 includes a fix for the first vulnerability. Unfortunately the second vulnerability still exists, and I hope to produce a patch for it soon enough. The vulnerability allows a user to read arbitrary files on the host system. In the meantime, this is not much of an issue for a properly containerised deployment. See the –sandbox argument for more information.
 Since then, I have created the Galaxy AWKScript wrapper.
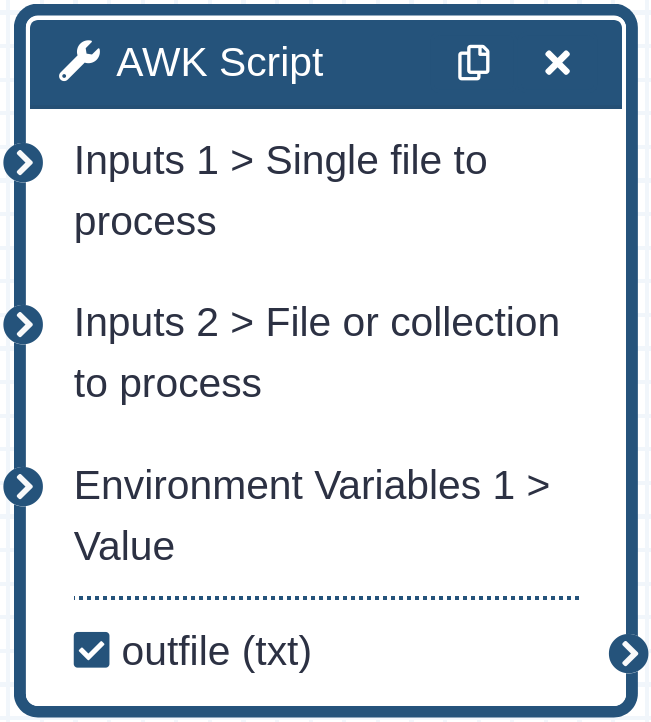
The wrapper forwards a number of very useful bits of information from the related Galaxy objects to gawk. Within the gawk
environment a variable ‘tool_input’ will be set to the index of the wrapper inputs, in order. You can combine this with
ARGIND to determine which file you are currently
operating on and its position in any possible input collection. A variable ‘tool_input_id’ is also set specifying the
current inputs dataset name or collection id. Beware that ARGIND will increment 3 between inputs as one is consumed
setting tool_input, another setting tool_input_id, and the third to specify the input file. Something to note is that
while Galaxy provides a dataset name to tool wrappers, it does not provide collection names.
Since then, I have created the Galaxy AWKScript wrapper.
The wrapper forwards a number of very useful bits of information from the related Galaxy objects to gawk. Within the gawk
environment a variable ‘tool_input’ will be set to the index of the wrapper inputs, in order. You can combine this with
ARGIND to determine which file you are currently
operating on and its position in any possible input collection. A variable ‘tool_input_id’ is also set specifying the
current inputs dataset name or collection id. Beware that ARGIND will increment 3 between inputs as one is consumed
setting tool_input, another setting tool_input_id, and the third to specify the input file. Something to note is that
while Galaxy provides a dataset name to tool wrappers, it does not provide collection names.
The following script demonstrates the information provided by the tool wrapper to gawk. The tool is provided with a collection input with three elements, and a single dataset input. Additionally, two environment variables were included.
BEGIN {
for (e in ENVIRON) {
print "ENVIRON["e"] = " ENVIRON[e];
}
print "tool_input", "ARGIND", "tool_input_id";
}
BEGINFILE {
print tool_input, ARGIND, tool_input_id;
nextfile;
}
 Output:
Output:
ENVIRON[env1] = val1
ENVIRON[env2] = val2
tool_input ARGIND tool_input_id
0 3 dataset1
0 5 dataset2
0 7 dataset3
1 10 single_dataset
As you can see, dataset1/2/3 are the collection element identifiers for the collection provided to input 0. single_dataset is the name of the dataset provided to input 1. env1/2 were provided to the script via the ENVIRON global. Download this demo workflow to test it yourself.
The environment inputs allow you to generalise your scripts, specifying constants with the tool invocation, or allow
attaching simple workflow parameter inputs.
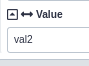 If you click the ⬌ icon on an Environment Variable value input, it will allow you to provide the value as a tool input
from a workflow Simple Parameter Input.
If you click the ⬌ icon on an Environment Variable value input, it will allow you to provide the value as a tool input
from a workflow Simple Parameter Input.
The wrapper inputs provide the ability to specify if they accept single inputs or if they accept collections of inputs. This is very useful when trying to get a specific collection mapping behavior. If you attach an optional workflow input to the input of this tool, the tool will run regardless of the input. If there is no input provided the BEGIN rule will be executed, but the BEGINFILE and subsequent rules will not.
This tool allows you to embed scripting logic within a workflow that is aware of the various properties of the workflow data. That data can be leveraged along with collection operation tools to provide a whole new level of functionality of Galaxy workflows.
Flow control
Galaxy does not (and rightfully so) provide an embedded scripting language for flow control. Remember, its workflows are nothing more than graph data structures. This can be frustrating when trying to create workflows that change their behavior based on input or intermediate data values. Combining my gawk tool wrapper, select collection operation tools, and “mapping over” functionality provides a robust mechanism for a variety of flow control operations.
This workflow demonstrates the simplest form of
flow control, a conditional:
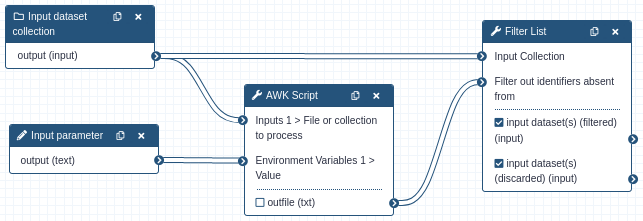 The gawk script contains:
The gawk script contains:
BEGIN {
if (ENVIRON['input'] != "yes") exit;
}
BEGINFILE {
print tool_input_id;
nextfile;
}
This will output each collection identifier to a text file for input as the filter criteria for the Filter List tool. Note that the nextfile command is an important optimisation, without it gawk will potentially iterate over every line of the datasets contents.
If the value provided by the workflow input parameter is anything other than ‘yes’, all elements of the collection are routed to the ‘discarded’ output of the Filter List step. This means that one output of the Filter List tool will be an empty collection, and any tool that is mapped over or assigned an empty collection also emits an empty collection without actually running the tool. This empty collection propagates throughout that branch of the workflow and is effectively not executed. Take care that some collection tools will continue to execute despite an empty collection, such as the Merge Collections tool.
The behaviour of this does not need to be bimodal, the script can contain any level of logic as to which datasets in the collection are filtered. You can even inspect the contents of the file and make decisions. The Filter List tool can be chained as many times as needed to get the level of branching required.
Collection of one
You may be thinking this is all great when working with collections of datasets, but most of your tools and workflows operate on single datasets. This all applies when you consider using a collection of size one. Your single dataset can happily be passed between tools that operate on the single dataset because the tool is mapped over when a collection is attached. All tools do not need to be mapped over in a workflow to allow for flow control. Only the branches and tools that could possibly receive a collection of size zero. The Build List, Merge Collection, and Extract Dataset tools can be used to enter and exit conditional portions of the workflow.
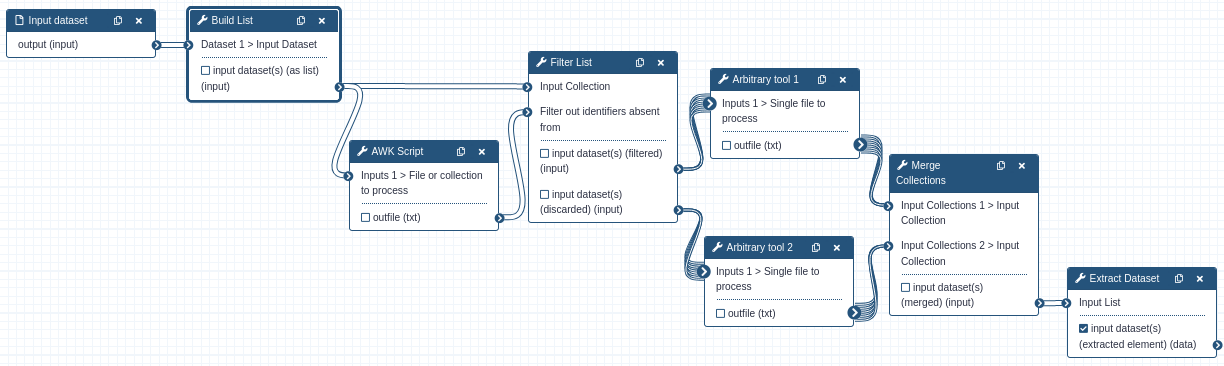 A single dataset is input into the Build List tool, which then outputs a collection of size one. The AWK script
evaluates the dataset and outputs which tool it wants the Filter List tool to route the dataset to. The collection
containing the output of whichever tool was executed is merged with the empty collection from the other tool. The
first (and only) dataset is then extracted from the collection as a single dataset again. Note that the Extract Dataset
tool will error if it receives an empty collection. You can only use this technique for a workflow that always produces
an output.
A single dataset is input into the Build List tool, which then outputs a collection of size one. The AWK script
evaluates the dataset and outputs which tool it wants the Filter List tool to route the dataset to. The collection
containing the output of whichever tool was executed is merged with the empty collection from the other tool. The
first (and only) dataset is then extracted from the collection as a single dataset again. Note that the Extract Dataset
tool will error if it receives an empty collection. You can only use this technique for a workflow that always produces
an output.
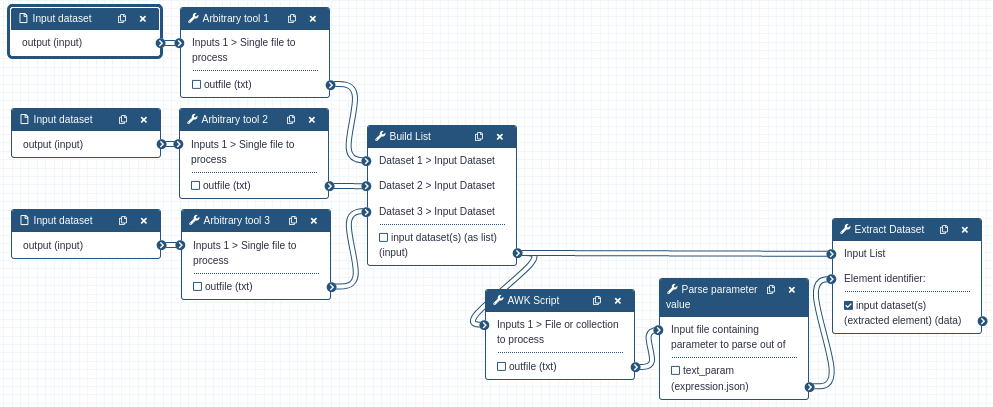 This is a different take on the use of the Extract Dataset tool. The datasets produced by each ‘Arbitrary tool’ can be
evaluated with the intent of picking the most desirable one. Nothing here is mapped over, the collection output from
the Build List tool is sent directly to the Extract Dataset tool. The Extract Dataset tool is set to extract the element
that is output by the AWK Script. The “Parse parameter value from dataset” tool is discussed in the next section.
This is a different take on the use of the Extract Dataset tool. The datasets produced by each ‘Arbitrary tool’ can be
evaluated with the intent of picking the most desirable one. Nothing here is mapped over, the collection output from
the Build List tool is sent directly to the Extract Dataset tool. The Extract Dataset tool is set to extract the element
that is output by the AWK Script. The “Parse parameter value from dataset” tool is discussed in the next section.
As you can see, manipulating collections with the output of the AWK Script tool can provide some powerful functionality. Many more combinations of the tools is possible, providing any functionality you might need.
Not limited to dataset inputs
The “Parse parameter value from dataset” tool is generally found under the Expression Tools section. It allows you to convert a dataset contents to a value that can be used for any tool parameter. Combining this with the AWKScript (or any other tool that can output a single value) allows you to specify tool parameters at runtime. To change a tool parameter to accept a value from the workflow, simply click the ⬌ icon above the input.
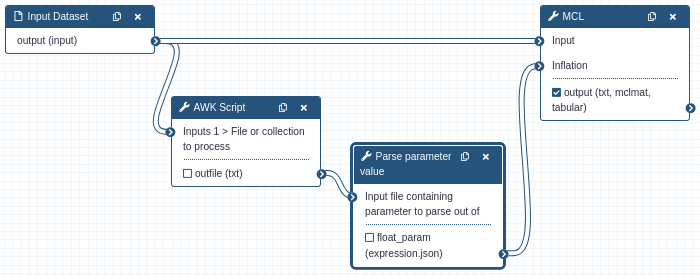 In this example, the input to a tool is first evaluated by the AWK Script to determine the “Inflation” value desired for
that dataset. The script outputs a number as a text dataset, that text is read in and parsed as a float and passed to the MCL tool.
In this example, the input to a tool is first evaluated by the AWK Script to determine the “Inflation” value desired for
that dataset. The script outputs a number as a text dataset, that text is read in and parsed as a float and passed to the MCL tool.
Apply rules to collection
So you have seen how you can manipulate flat collections for a variety of purposes, and you can generate those collections using the Build List tool. What if you want to generate some of the more complex collection types to take advantage of the advanced mapping properties previously mentioned? This is where the “Apply rules to collection” tool comes in.
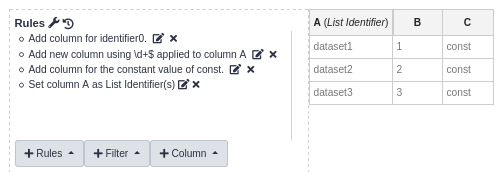
It is a tool that allows you to rework an existing collection into a completely different collection structure. Take special note of its “Split Columns” rule, along with the “Regular Expression”, and “Concatenate Columns” column creation options. They can be leveraged against the way the Flatten Collection tool generates collection element identifiers for some useful transformations.
 An easily overlooked feature of this tool is its JSON view of the rules. Click the 🔧 to open it. This allows you to quickly
copy/paste a rule set into and out of the tool for reuse. If you plan to use this tool in a workflow, I recommend you
manually generate a input collection and build the rules using its dynamic preview feature. This only appears when viewing
the tool outside of the workflow editor. Once you are done generating the rules, open the JSON view, copy them out, and
paste into the JSON view of the tool inserted in your workflow.
An easily overlooked feature of this tool is its JSON view of the rules. Click the 🔧 to open it. This allows you to quickly
copy/paste a rule set into and out of the tool for reuse. If you plan to use this tool in a workflow, I recommend you
manually generate a input collection and build the rules using its dynamic preview feature. This only appears when viewing
the tool outside of the workflow editor. Once you are done generating the rules, open the JSON view, copy them out, and
paste into the JSON view of the tool inserted in your workflow.
Copy and paste this example to demonstrate how to generate a list:list from a list. This can be useful when you want to map over a tool that already accepts a collection.
A more interesting rule set takes a list of datasets (A,B,C,D) and produces a list:list of those datasets in a pairwise combination ([A,B],[B,C],[C,D]). If a tool accepts a collection, it will be mapped over this list:list, where each invocation receives a pair. This is different from the way a list:pair collection works. A list:pair generally wants to evaluate as ([A,B],[C,D]) where each pair has an exclusive relationship between the datasets.
Writing your own tool wrappers
You may find yourself wanting to write a Galaxy tool wrapper for your favorite tool. Unfortunately the environment and values provided to the tool command template are not well documented. I have produced a tool that will display much of the execution environment available to a tool command template. The tool needs to be manually installed as it is far too dangerous of a tool to distribute via the toolshed. DO NOT INSTALL THIS TOOL ON A PUBLIC SERVER, it will output all the critical secrets of the Galaxy deployment and system. This tool also does everything that a tool wrapper should never do and will likely need to be tweaked depending on the version of Galaxy it is installed to before it will run.
Be sure to check out Galaxy’s documentation for the tool XML file as your starting place when developing a wrapper. It isn’t well advertised in the Galaxy documentation, but a tool wrapper command is compiled using the Cheetah Template Engine and all the engines features are available.
Conclusion
Hopefully by this point you have everything you need to start creating robust, well-formed pipelines. There are a million more tiny little features scattered about Galaxy that can be quite useful, and I encourage you to explore.
It is important to point out that many of the techniques mentioned here will fail if used in a sub-workflow. This is due to a bug in earlier releases of Galaxy. I have worked with the Galaxy developers to resolve these issues in version 20.09 and later versions.
Check out the Awesome Galaxy repository for a community maintained list of awesome Galaxy resources.
If you need help with a tool or specific use case of Galaxy, I recommend you visit the official help forum. There is also a friendly community of Galaxy users and developers hanging out in the Galaxy Gitter.

Leave a Comment